Protein Engineering
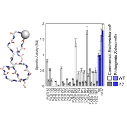
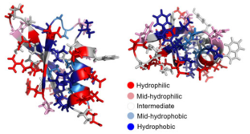
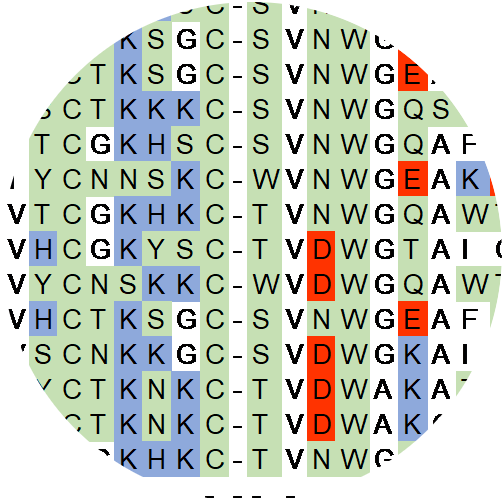
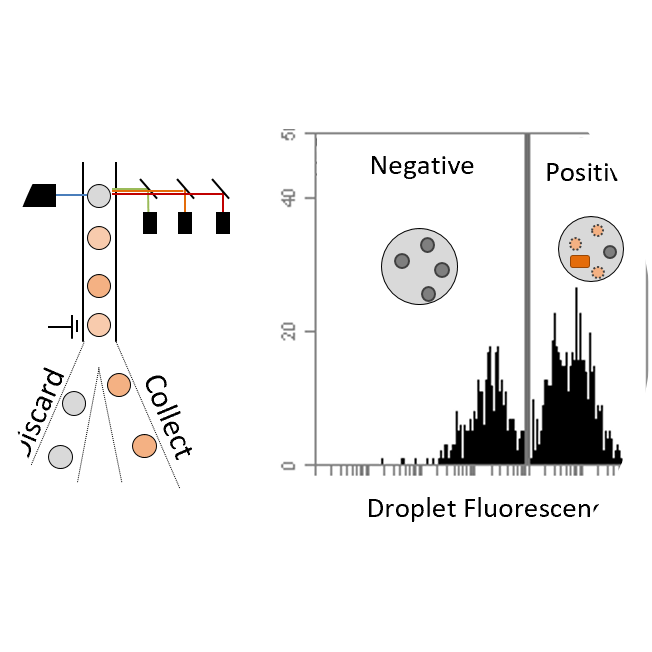
We engineer proteins with novel functions using, predominantly, yeast surface display and directed evolution. The relationships amongst protein sequence and function are studied to identify functional scaffolds for de novo discovery and design combinatorial libraries that more efficiently search the enormity of protein sequence space. We develop technologies for enhanced selection of multiple phenotypes (binding, antimicrobial activity, catalysis) and diversification of lead molecules to enable more effective evolution.
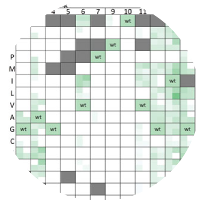
Combinatorial Scaffold and Library Design
Selection Technologies




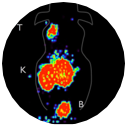
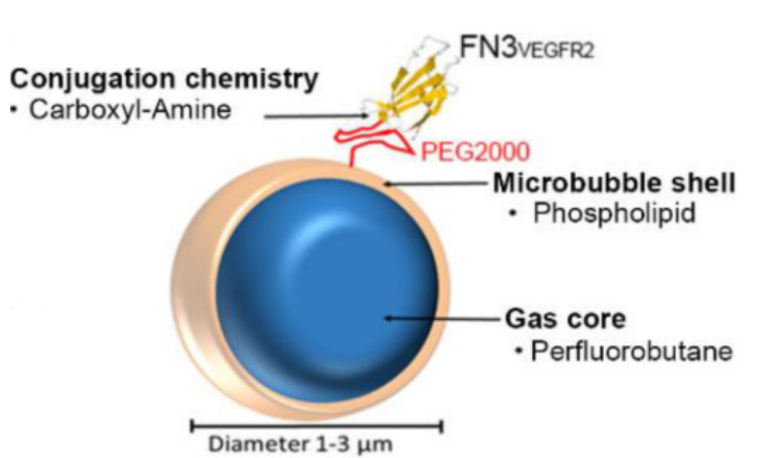
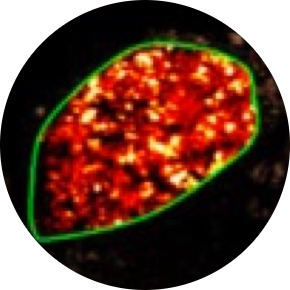
Diagnostics and Therapeutics
Multifunctional Proteins