Publications
|
|
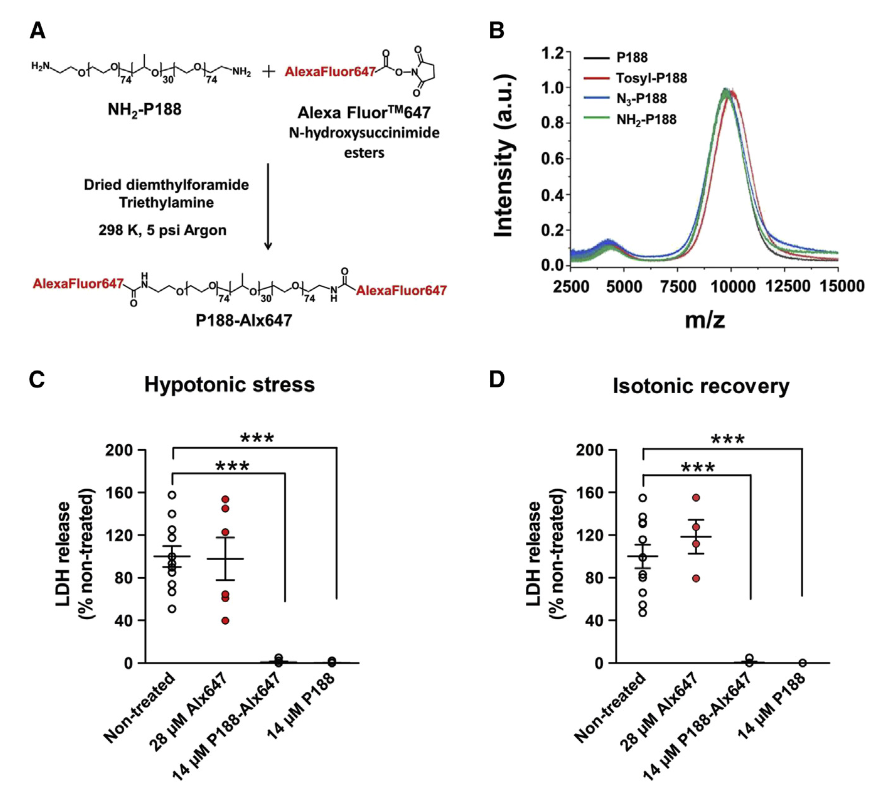
Angulski, A., Cohen, H., Kim, M., Hahn, D., Van Zee, N., Lodge, T., Hillmyer, M., Hackel, B.J., Bates, F., Metzger, J. "Molecular homing and retention of muscle membrane stabilizing copolymers by non-invasive optical imaging in vivo" Molecular Therapy: Methods and Clinical Development 2023.
|
|
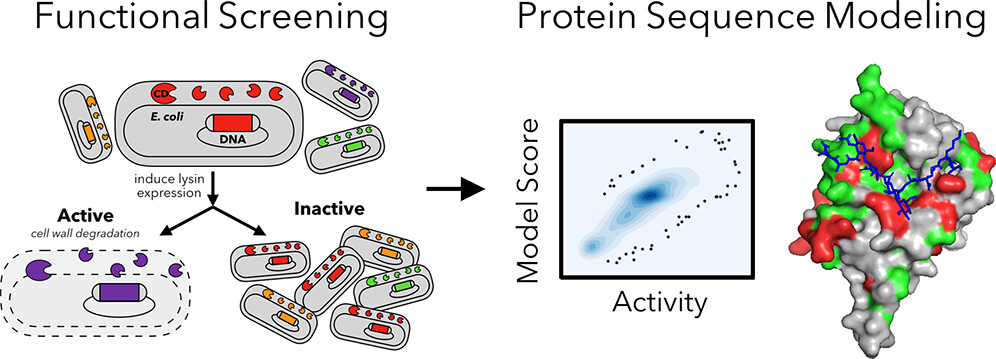
Tresnak, D., Hackel, B.J. "Deep Antimicrobial Activity and Stability Analysis Inform Lysin Sequence-Function Mapping" ACS Synthetic Biology 2023.
|
|
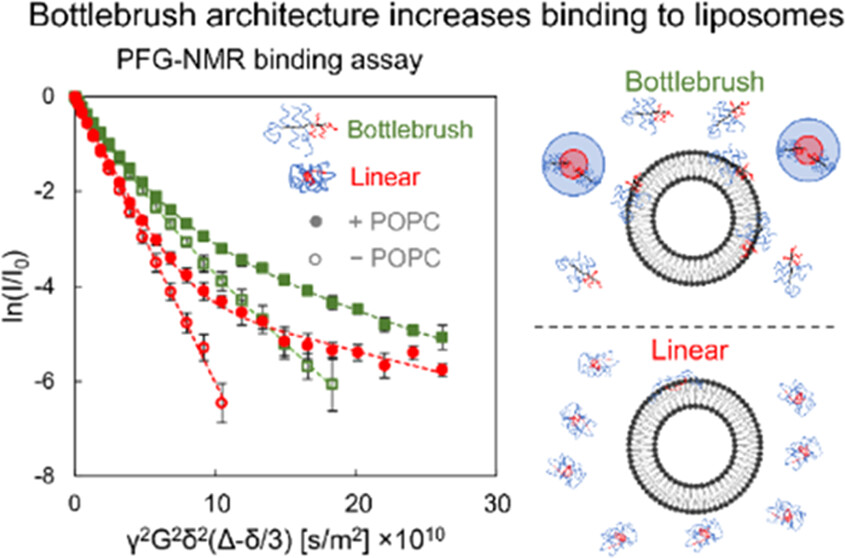
Hassler, J.F., Crabtree, A., Liberman, L., Bates, F., Hackel, B.J., Lodge, T. "Effect of Bottlebrush Poloxamer Architecture on Binding to Liposomes" Biomacromolecules 2022.
|
|
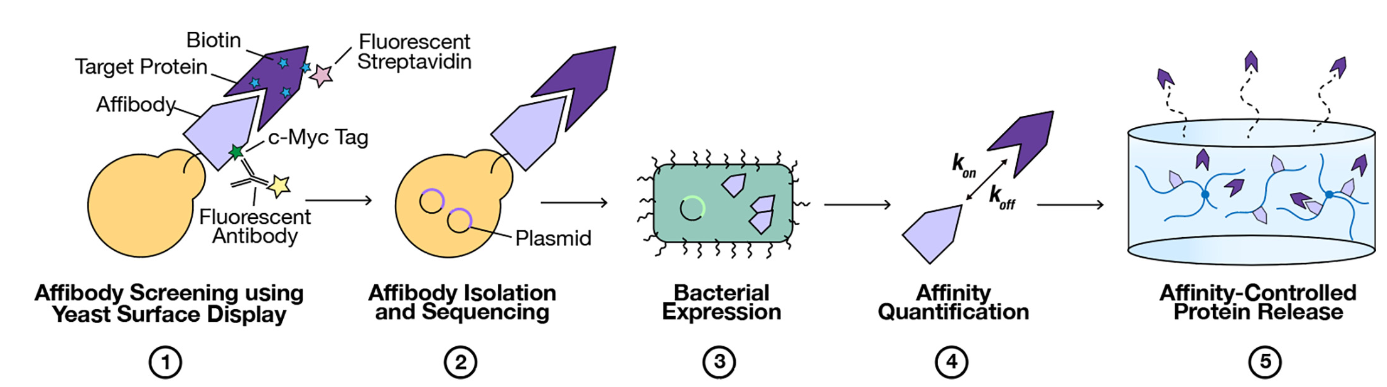
Bostock, C., Teal, C., Dang, M., Golinski, A.W., Hackel, B.J., Shoichet, M. "Affibody-mediated controlled release of fibroblast growth factor 2" Journal of Controlled Release 2022.
|
|
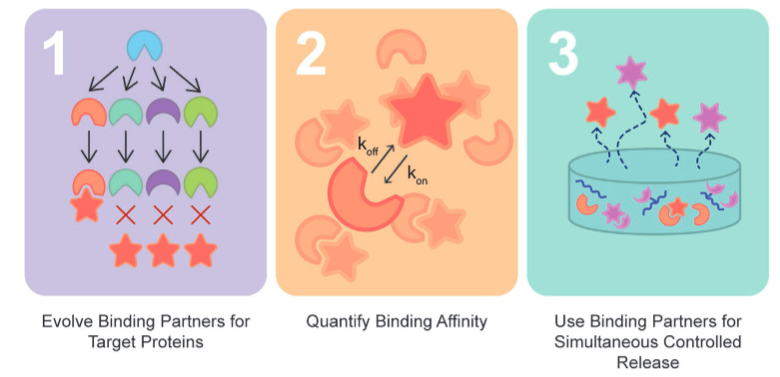
Teal, C., Hettiaratchi, M., Ho, M., Ortin-Martinez, A., Ganesh, A., Pickering, A., Golinski, A.W., Hackel, B.J., Wallace, V., Shoichet, M. "Directed Evolution Enables Simultaneous Controlled Release of Multiple Therapeutic Proteins from Biopolymer-Based Hydrogels" Advanced Materials 2022.
|
|
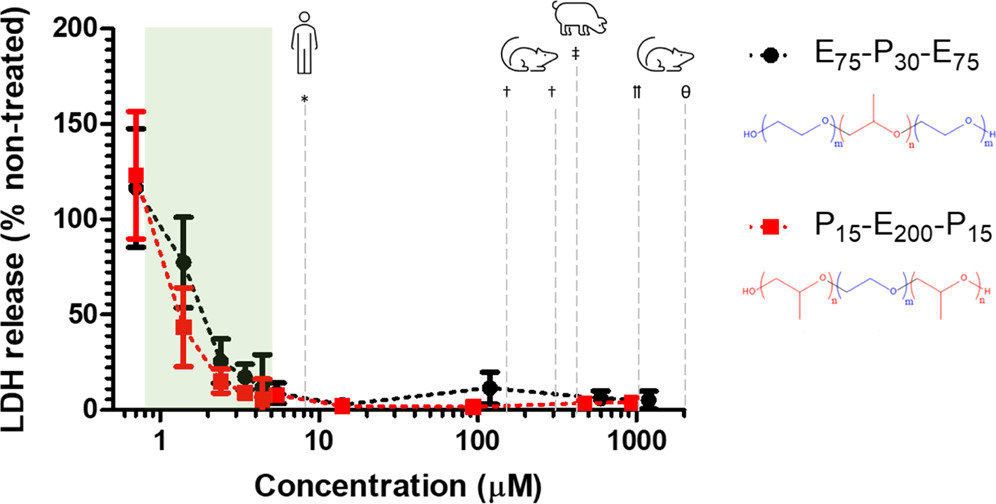
Crabtree, A., Bates, F., Hackel, B.J. "Concentration Threshold for Membrance Protection by PEO-PPO Block Copolyjers with Variable Molecular Architectures" ACS Applied Polymer Materials 2022.
|
|
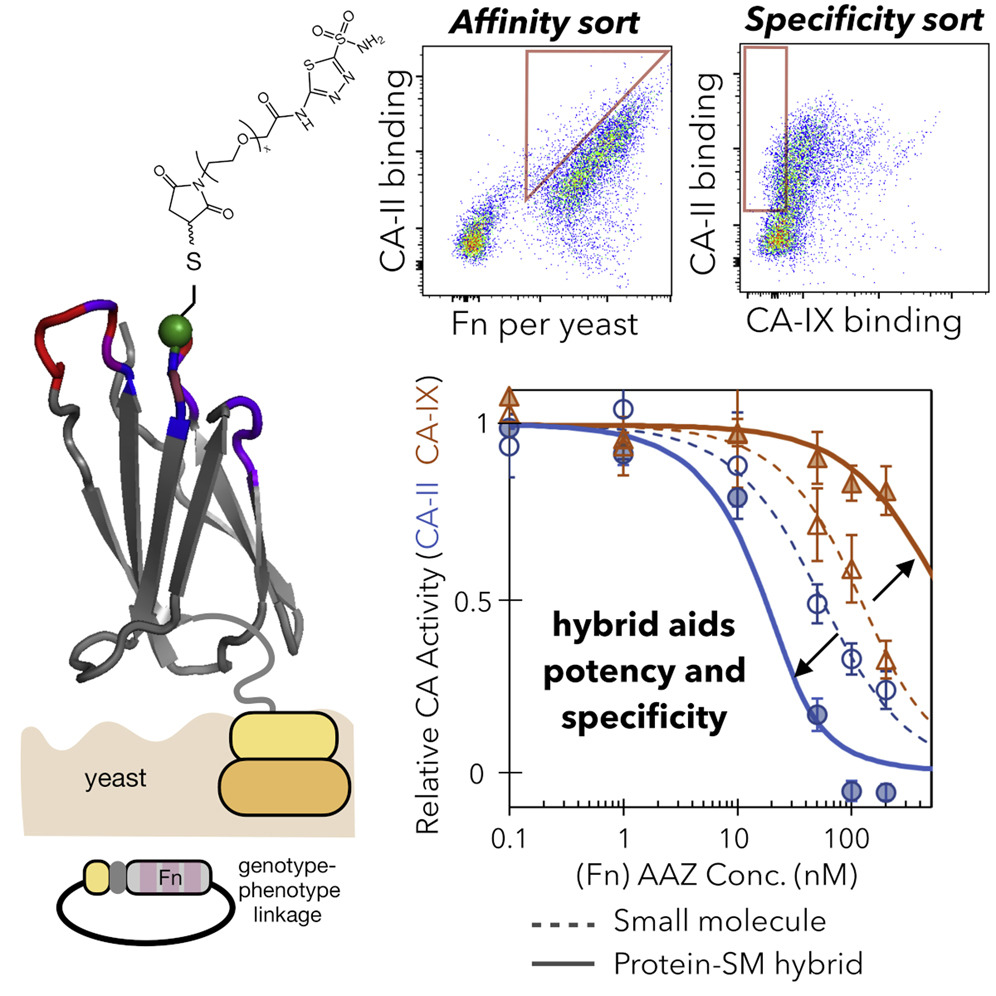
Lewis, A.K., Harthorn, A., Johnson S.M., Lobb, R.R., Hackel, B.J. "Engineered protein-small molecule conjugates empower selective enzyme inhibition" Cell Chemical Biology 2022.
|
|
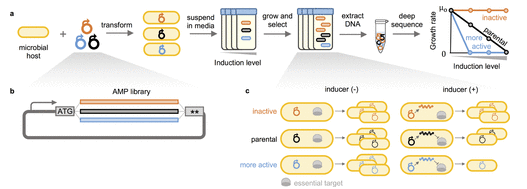
DeJong, M., Ritter, S., Fransen, K., Tresnak, D., Golinski, A., Hackel, B.J. "A Platform for Deep Sequence-Activity Mapping and Engineering Antimicrobial Peptides" ACS Synthetic Biology 2021.
|
|
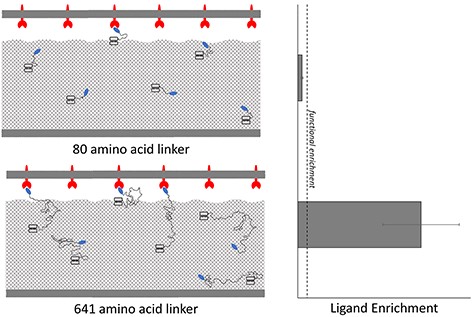
Lown, P.S., Cai, J., Ritter, S., Otolski, J., Wong, R., Hackel, B.J. "Extended yeast surface display linkers enhance the enrichment of ligands in direct mammalian cell selections" Protein Engineering Design and Selection 2021.
|
|
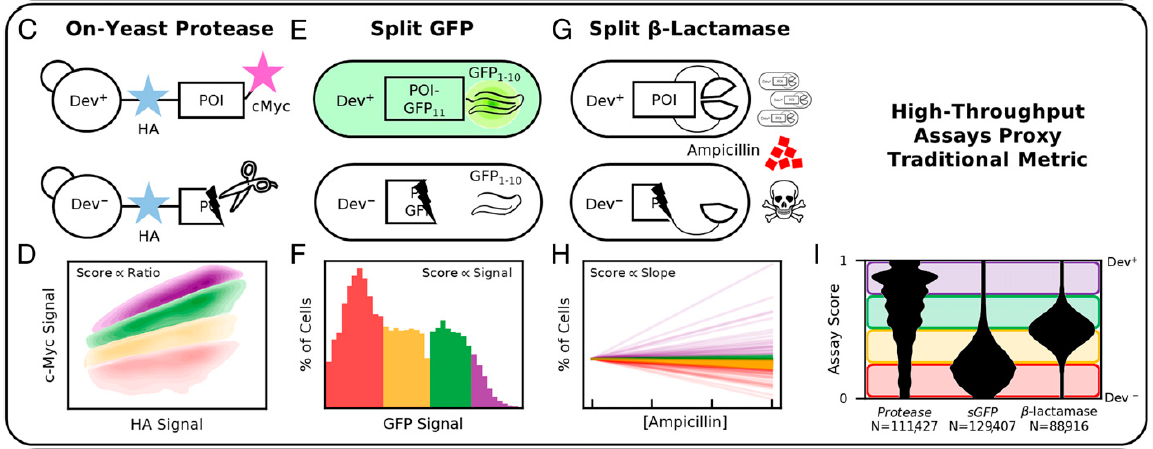
Golinski, A., Mischler, K., Laxminarayan, S., Neurock, N., Fossing, M., Pichman, H., Martiniani, S., Hackel, B.J. "High-throughput developability assays
enable library-scale identification of producible protein scaffold variants" PNAS Applied Biological Sciences 2021.
|
|
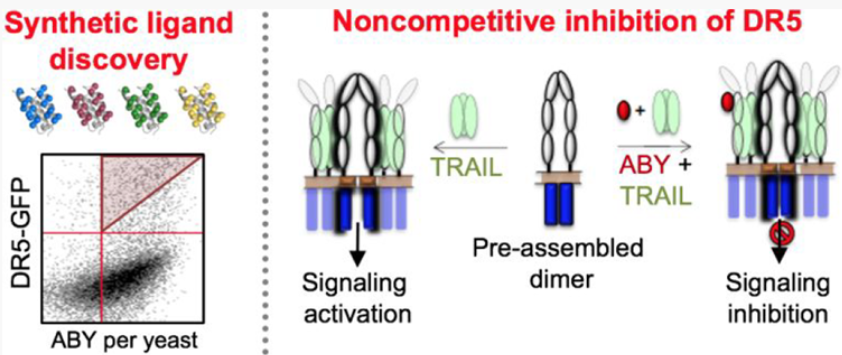
Vunnam, N., Szymonski, S., Hirsova, P., Gores, G.J., Sachs, J.N., Hackel, B.J. "Noncompetitive Allosteric Antagonism of Death Receptor 5 by a Synthetic Affibody Ligand" ACS Biochemistry 2020.
|
|
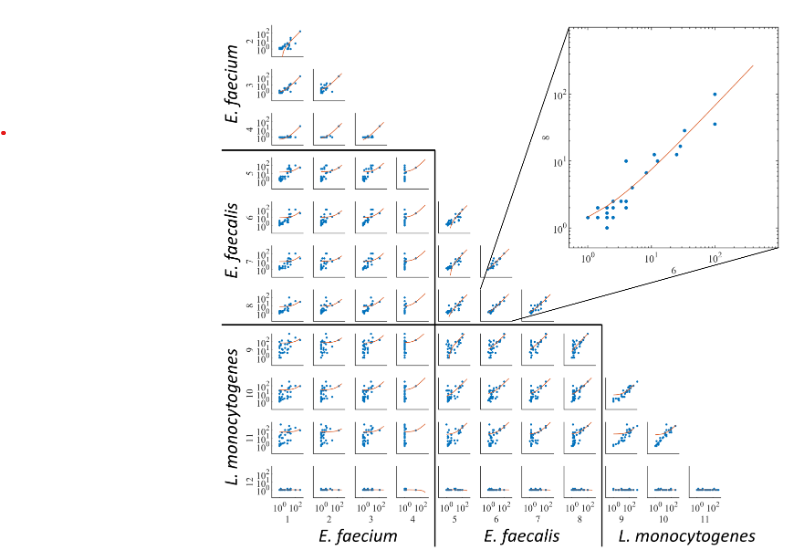
Tresnak, D.T., Hackel, B.J. "Mining and statistical modeling of natural and variant class IIa bacteriocins elucidates activity and selectivity profiles across species" Appl. Environ. Microbiol. 2020.
|
|
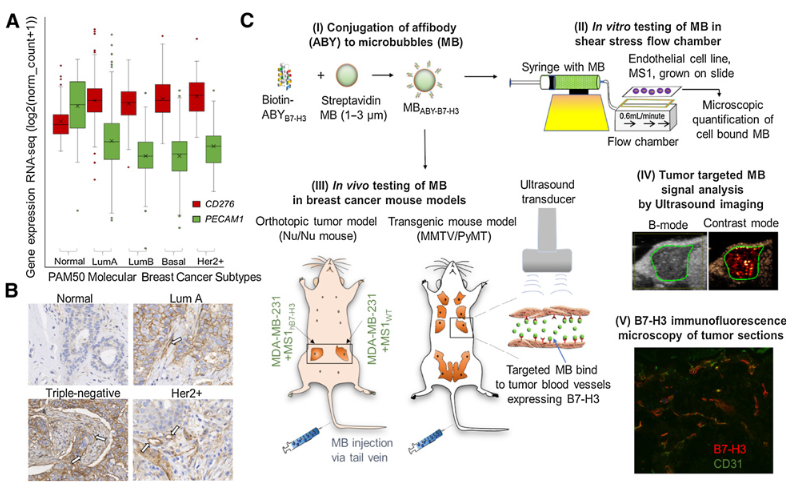
Bam, R., Lown, P.S., Stern, L.A., Sharma, K., Wilson, K.E., Bean, G.R., Lutz, A.M., Paulmurugan, R., Hackel, B.J., Dahl, J., Abou-Elkacem, L. "Efficacy of Affibody-Based Ultrasound Molecular Imaging of Vascular B7-H3 for Breast Cancer Detection" Clinical Cancer Research 2020.
|
|
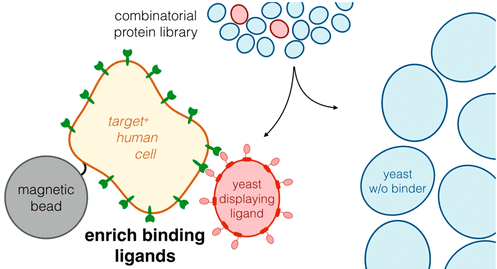
Lown, P.S., Hackel, B.J. "Magnetic Bead-Immobilized Mammalian Cells Are Effective Targets to Enrich Ligand-Displaying Yeast" ACS Combinatorial Science 2020.
|
|
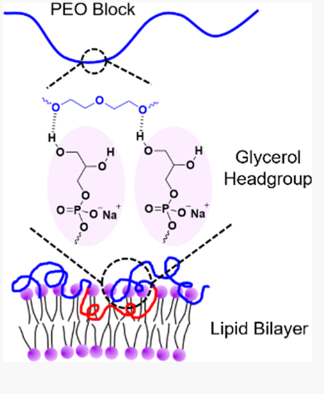
Zhang, W., Metzger, J.M., Hackel, B.J., Bates, F., Lodge, T.P. "Influence of the Headgroup on the Interaction of Poly(ethyleneoxide)-Poly(propylene oxide) Block Copolymers with Lipid Bilayers" J. PHys. Chem B 2020.
|
|
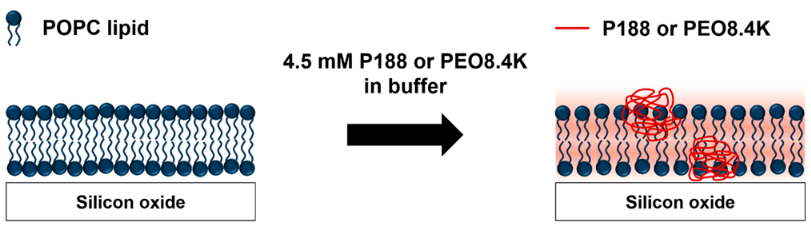
Kim, M., Heinrich, F., Haugstad, G., Yu, G., Yuan, G., Satija, S.K., Zhang, W., Seo, H.S., Metzger, J.M., Azarin, S.M., Lodge, T.P., Hackel, B.J., Bates, F.S. "Spatial Distribution of PEO-PPO-PEO Block Copolymer and PEO Homopolymer in Lipid Bilayers" Langmuir 2020.
|
|
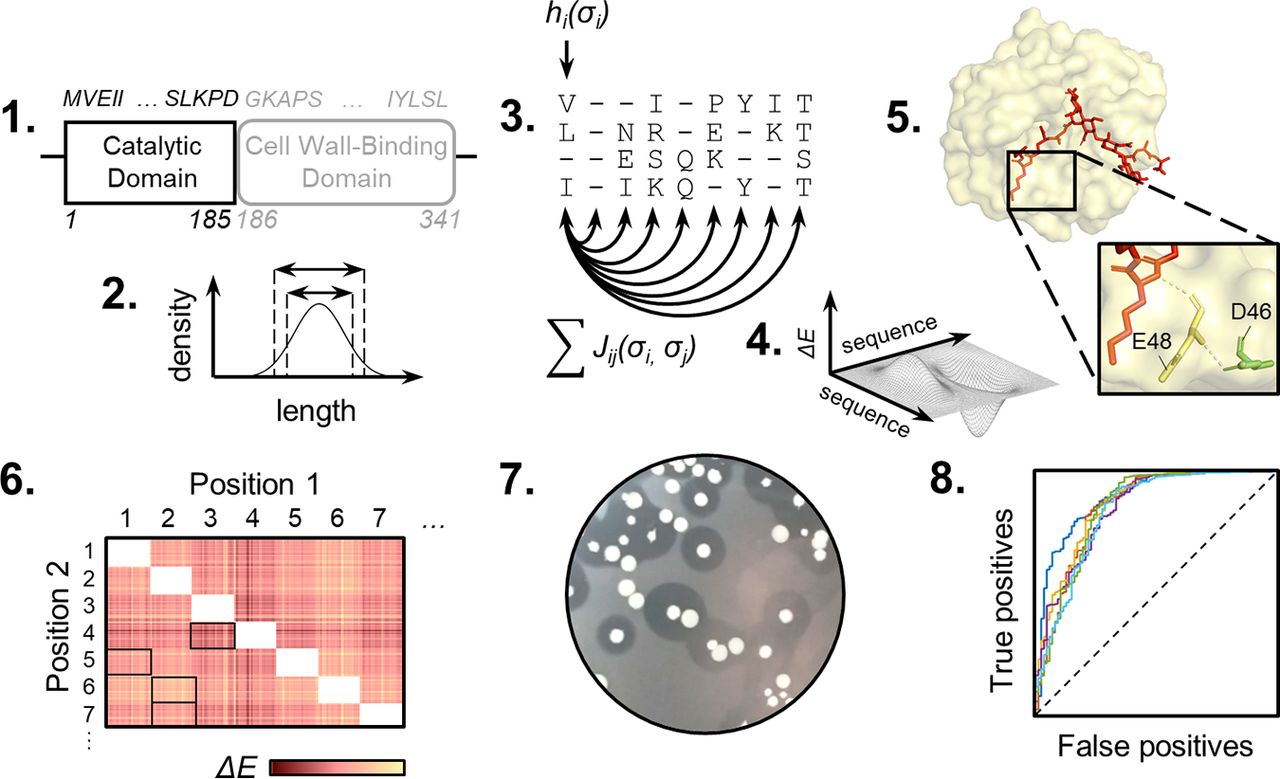
Baryakova, T.H., Ritter, S.C., Tresnak, D.T., Hackel, B.J. "Computationally Aided Discovery of LysEFm5 Variants with Improved Catalytic Activity and Stability" App. & Env. Microbio. 2020.
|
|
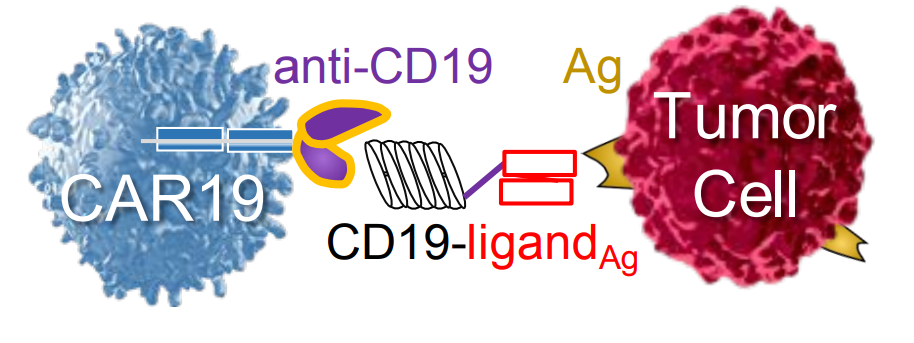
Klesmith, J.R., Su, L., Wu, L., Schrack, I.A., Dufort, F.J., Birt, A.,
Ambrose, C., Hackel, B.J., Lobb, R.R., Rennert, P.D. "Retargeting CD19
CAR T cells via engineered CD19-fusion proteins" Mol. Pharm. 2019.
|
|
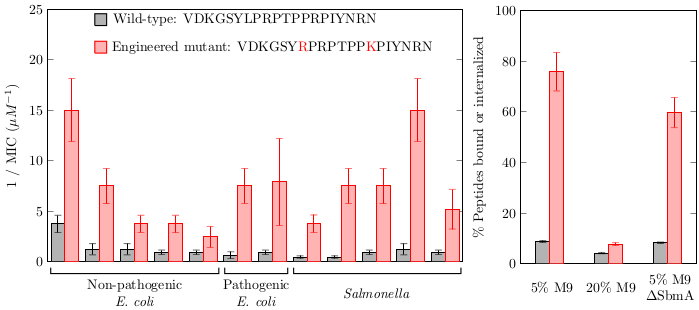
Lai, P.K., Tresnak, D.T., Hackel, B.J., "Identification and elucidation of
proline-rich antimicrobial peptides with enhanced potency and delivery",
Biotechnol. Bioeng. 2019.
|
|
Ritter, S.C., Hackel, B.J.,
"Validation and stabilization of a prophage lysin of Clostridium perfringens
by yeast surface display and co-evolutionary models", App. & Exp. Microbio. 2019.
|
|
Golinski, A.W., Holec, P.V., Mishler, K.M., Hackel, B.J.,
"Biophysical Characterization Platform Informs Protein Scaffold Evolvability",
ACS Combinatorial Science 2019.
|
|
Stern, L.A., Lown, P., Kobe, A., Abou-Elkacem, L. Willmann, J.K., and
Hackel, B.J., "Cellular-Based Selections Aid Yeast-Display Discovery of
Genuine Cell-Binding Ligands: Targeting Oncology Vascular Biomarker
CD276", ACS Combinatorial Science 2019.
|
|
Klesmith, J.R., Hackel, B.J.,
"Improved mutant function prediction via PACT: Protein Analysis and Classifier Toolkit"
Bioinformatics. 2018.
|
|
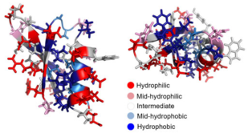
Du, F., Kruziki, M.A., Zudock, E.J., Zhang, Y., Lown, P.S., Hackel, B.J.,
"Engineering an EGFR-binding Gp2 domain for increased hydrophilicity"
Biotechnol. Bioeng. 2018.
|
|
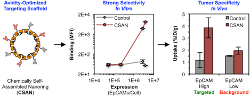
Csizmar, C.M., Petersburg, J.R., Perry, T.J., Rozumalski, L., Hackel, B.J., Wagner, C.R.,
"Multivalent Ligand Binding to Cell Membrane Antigens: Defining the
Interplay of Affinity, Valency, and Expression Density" J. Am. Chem. Soc. 2018.
|
|
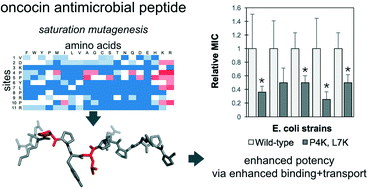
Lai, P.K., Geldart, K., Ritter, S., Kaznessis, Y.N., Hackel, B.J.,
"Systematic mutagenesis of oncocin reveals enhanced activity and
insights into the mechanisms of antimicrobial activity" Mol. Sys. Des. & Eng. 2018 6: 930-941.
|
|
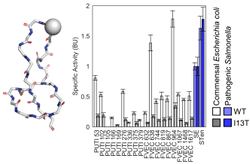
Ritter, S.C., Yang, M.L., Kaznessis Y.N., Hackel B.J.,
"Multispecies activity screening of microcin J25 mutants yields
antimicrobials with increased specificity toward pathogenic Salmonella
species relative to human commensal Escherichia coli" Biotechnol. Bioeng.
2018 115: 2394-2404.
|
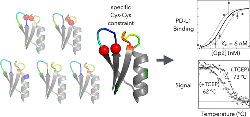
|
Kruziki, M.A., Sarma, V., Hackel, B.J., "Constrained Combinatorial
Libraries of Gp2 Proteins Enhance Discovery of PD-L1 Binders" ACS Comb.
Sci. 2018 20: 423-435.
|
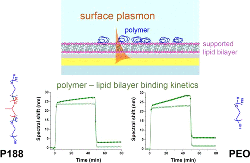
|
Kim, M., Vala, M., Ertsgaard, C.T., Oh, S., Lodge, T.P., Bates, F.S., Hackel, B.J.,
"Surface Plasmon Resonance Study of the Binding of PEO-PPO-PEO
Triblock Copolymer and PEO Homopolymer to Supported Lipid Bilayers"
Langmuir. 2018 34: 6703-6712.
|

|
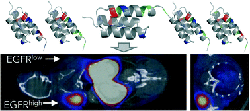
Case, B.A., Kruziki, M.A., Johnson, S.M., and Hackel, B.J.,
"Engineered charge redistribution of Gp2 proteins through guided
diversity for improved PET imaging of epidermal growth factor
receptor" Bioconj. Chem. 2018 29: 1646-1658.
|

|
Csizmar C.M., Petersburg J. Hendricks A., Stern L.A., Hackel B.J.,
and Wagner CR, "Engineering reversible cell-cell interactions with
lipid anchored prosthetic receptors" Bioconj. Chem. 2018 29: 1291-1301.
|
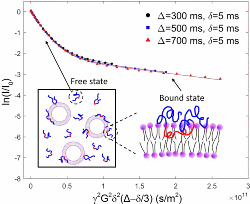
|
Zhang, W., Haman, K.J., Metzger, J.M., Hackel, B.J., Bates, F.S.,
and Lodge, T.P. "Quantifying Binding of Ethylene Oxide-Propylene Oxide
Block Copolymers with Lipid Bilayers" Langmuir. 2017 33: 12624-12634.
|
|
Case, B.A. and Kruziki, M.A. and Stern, L.A. and Hackel, B.J.
"Evaluation of affibody charge modification identified by synthetic
consensus design in molecular PET imaging of epidermal growth factor receptor"
Mol. Syst. Des. Eng. 2018 3: 171-182.
|
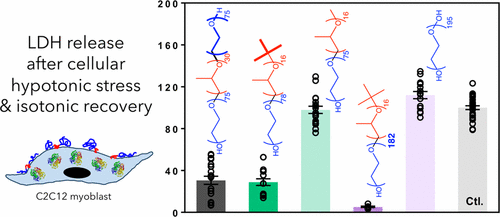
|
Kim, M., Haman, K.J., Houang, E.M., Zhang, W., Yannopoulos, D., Metzger, J.M.,
Bates, F.S., Hackel, B.
"PEO-PPO Diblock Copolymers Protect Myoblasts from Hypo-Osmotic
Stress In Vitro Dependent on Copolymer Size, Composition, and Architecture"
Biomacromolecules. 2017 18: 2090-2101.
|
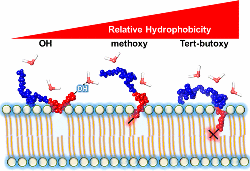
|
Houang, E.M., Haman, K.J., Kim, M., Zhang, W. Lowe, D.A., Sham, Y.Y., Lodge,
T.P., Hackel, B.J., Bates, F.S., and Metzger, J.M.,
"Chemical end group modified diblock copolymers elucidate anchor
and chain mechanism of membrane stabilization" Mol. Pharm. 2017 14:
2333-2339.
|
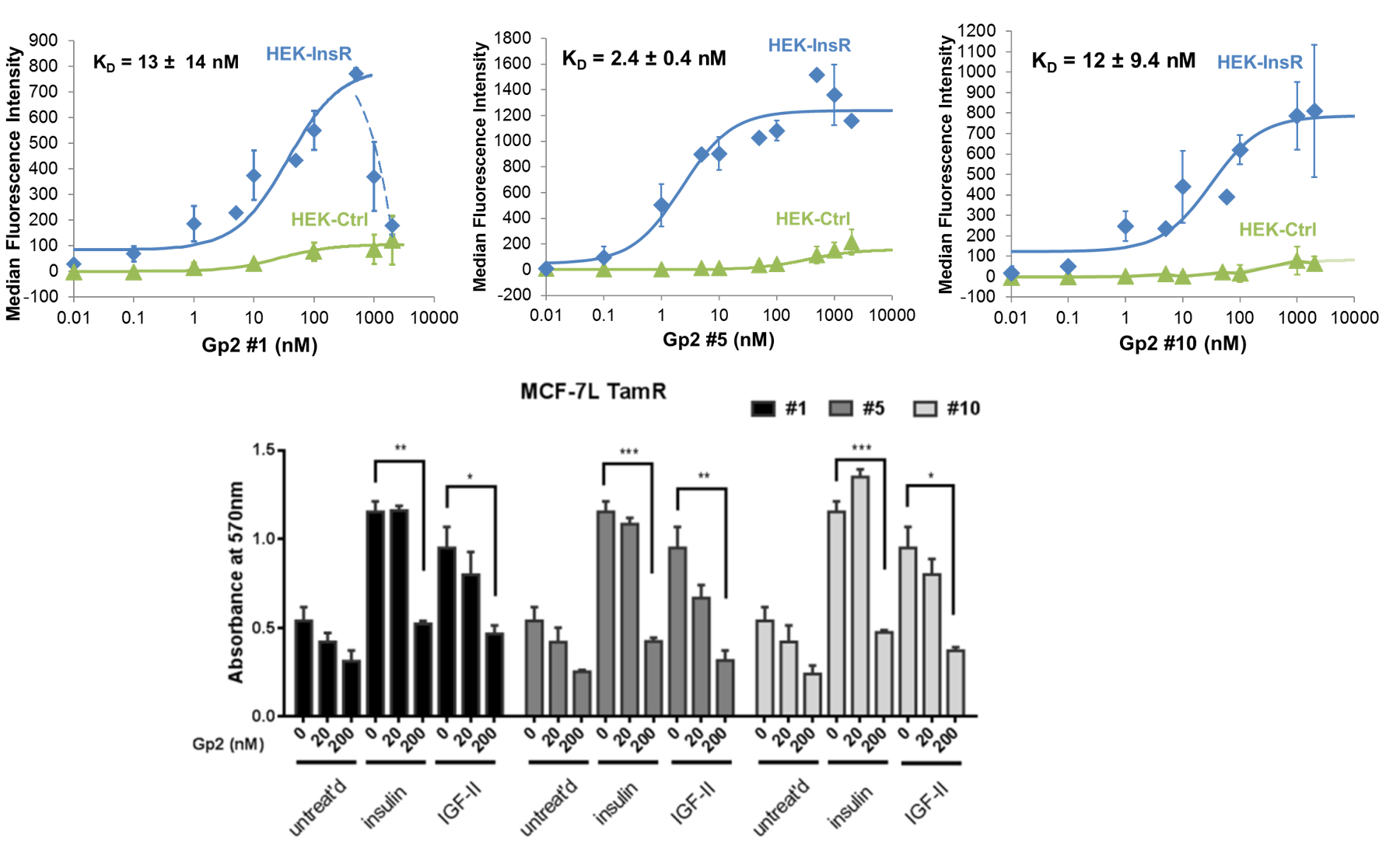
|
Chan, J.Y., Hackel, B., Yee, D.
"Targeting insulin receptor in breast cancer using small engineered protein scaffolds"
Mol. Cancer. Ther. 2017 16: 1324-1334.
|
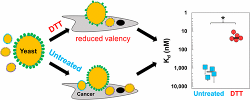
|
Stern, L., Csizmar, C., Woldring, D., Wagner, C., Hackel, B.
"Titratable Avidity Reduction Enhances
Affinity Discrimination in Mammalian Cellular Selections of Yeast-Displayed Ligands"
ACS. Comb. Sci. 2017 19: 315-323.
|
|
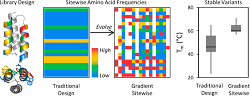
Woldring, D., Holec, P., Stern, L., Du, Y., Hackel, B.
"A Gradient of Sitewise Diversity Promotes Evolutionary
Fitness for Binder Discovery in a Three-Helix Bundle Protein Scaffold"
Biochem. 2017 56: 1656-1671.
|
|
Johnson, S., Javner, C., Hackel, B. "Development and
implementation of a protein-protein binding experiment to teach intermolecular
interactions in high school or undergraduate classrooms"
J. Chem. Ed. 2017 94: 367-374.
|
|
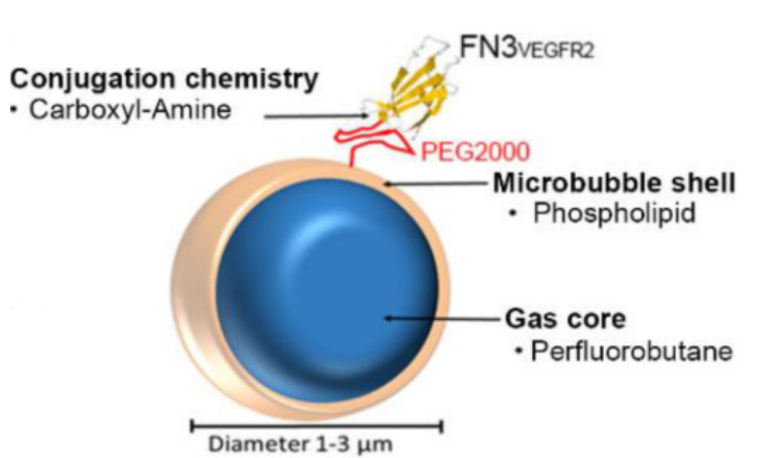
Abou-Elkacem, L., Wilson, K.E., Johnson, S.M., Chowdhury, S.M., Bachawal, S.,
Hackel, B.J., Tian, L., Willmann, J.K., "Ultrasound Molecular Imaging of the Breast Cancer
Neovasculature using Engineered Fibronectin Scaffold Ligands: A Novel Class of
Targeted Contrast Ultrasound Agent" Theranostics. 2016 6(11): 1740-1752.
|
|
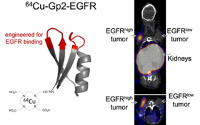
Kruziki, M., Case, B., Chan, J., Zudock, E., Woldring, D., Yee, D., and Hackel, B.J.,
"64Cu-Labeled Gp2 Domain for PET Imaging of Epidermal Growth Factor Receptor"
Mol. Pharm. 2016 13: 3747-3755.
|
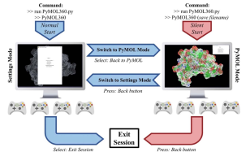
|
Holec, P.V., and Hackel, B.J., "PyMOL360: Multi-user gamepad control of molecular
visualization software" J. Comput. Chem. 2016 37(30) 2667-69.
|
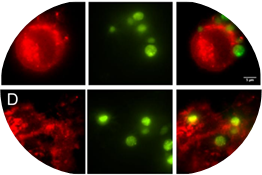
|
Stern, L.A., Schrack, I.A., Johnson, S.M., Deshpande, A.,
Bennett, N.R., Harasymiw, L.A., Gardner, M.K., Hackel, B.J.,
"Geometry and expression enhance enrichment of functional yeast-displayed
ligands via cell panning." Biotechnol. Bioeng. 2016 113:2328-41.
|
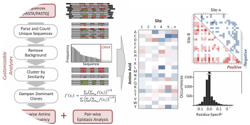
|
Woldring, D.R., Holec, P.V., and Hackel, B.J., "ScaffoldSeq: Software for characterization
of directed evolution populations" Proteins. 2016 84:869-74.
|
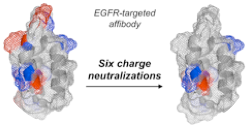
|
Case, B.A., and Hackel, B.J., "Synthetic and natural consensus design for engineering charge
within an affibody targeting epidermal growth factor receptor" Biotechnol. Bioeng.
2016 113:1628-38.
|

|
Woldring, D.R., Holec, P.V., Zhou, H., and Hackel, B.J., "High-throughput ligand discovery
reveals a sitewise gradient of diversity in broadly evolved hydrophilic fibronectin domains"
PLOS One. 2015, 10, e0138956.
|
|
McClintock, M., Kaznessis, Y.N., and Hackel B.J., "Enterocin A mutants identified by saturation
mutagenesis enhance potency towards vancomycin-resistant Enterococci" Biotechnol. Bioeng. 2015.
|
|
Kruziki, M.A., Bhatnagar, S., Woldring, D.R., Duong, V.T., and Hackel B.J., "A 45-amino
acid scaffold mined from the Protein Data Bank for high affinity ligand engineering" Chem. Biol.
2015 22:946-956.
|
|
Morgounova, E., Johnson, S.M., Shao, Q., Hackel, B.J., and Ashkenazi, S., "Lifetime-based
photoacoustic probe activation modeled by a dual methylene blue-lysine conjugate"
Progress in Biomedical Optics and Imaging - Proceedings of SPIE. 2014;8943.
|
|
Burns, M.L., Malott, T.M., Metcalf, K.J., Hackel, B.J., Chan, J.R., and Shusta, E.V.,
"Directed evolution of brain-derived neurotrophic factor for improved folding and expression
in yeast" Appl. Env. Microb. 2014 80, 5732-5742.
|
|
Hackel, B.J., "Ligand engineering using yeast surface display" Methods Mol. Biol.
2014, 1163, 257-271.
|
|
Natarajan, A., Hackel, B.J., and Gambhir, S.S., "A novel engineered anti-CD20 tracer enables
early time PET imaging in a humanized transgenic mouse model of B-cell non-Hodgkins lymphoma"
Clin. Cancer Res. 2013 19, 6820-6829.
|
|
Stern, L.A., Case, B.A., and Hackel, B.J., "Alternative non-antibody protein scaffolds
for molecular imaging of cancer" Curr. Opin. Chem. Engr. 2013 2, 425-432.
|
|
Morgounova, E., Shao, Q., Hackel, B.J., Thomas, D., and Ashkenazi, S., "Photoacoustic
lifetime contrast between methylene blue monomers and statically quenched dimers as a model
for dual-labeled activatable probes" J. Biomed. Opt. 2013 5:56004.
|
|
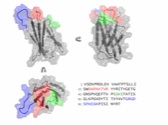
Chen, T.F., de Picciotto, S., Hackel, B.J., and Wittrup, K.D., "Engineering
fibronectin-based binding proteins by yeast surface display" in
Meth. Enzymol., Methods in Protein Design, (Keating, A., ed) 2013 523:303-326.
|
|
Hackel, B.J., Kimura, R.H., Miao, Z., Liu, H., Sathirachinda, A., Cheng, Z.,
Chin, F.T., and Gambhir, S.S., "18F-Labeled Cystine Knot Peptides for PET
Imaging of Integrin αvβ6" J. Nucl. Med. 2013 54:1101-1105.
|
|
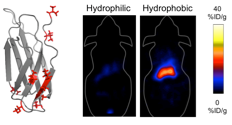
Hackel, B.J., Sathirachinda, A., and Gambhir, S.S., "Designed hydrophilic and
charge mutations of the fibronectin domain: towards tailored protein biodistribution"
Prot. Engr. Des. Sel. 2012 25:639-648.
|
|
|
Hackel, B.J., Kimura, R.H., and Gambhir, S.S., "Use of 64Cu-labeled Fibronectin Domain
with EGFR-Overexpressing Tumor Xenograft: Molecular Imaging"
Radiology. 2012:179-188.
|
|
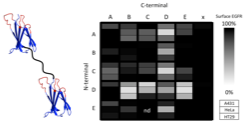
Hackel, B.J., Neil, J.R., White, F.M., and Wittrup, K.D., "Epidermal growth factor
receptor downregulation by small heterodimeric binding proteins" Prot. Engr. Des. Sel. 2012 25, 47-57.
|
|
Kimura, R.H., Teed, R., Hackel, B.J., Pysz, M.A., Chuang, C.Z., Sathirachinda, A., et al.,
"Pharmacokinetically Stabilized Cystine Knot Peptides That Bind Alpha-v-Beta-6 Integrin
with Single-Digit Nanomolar Affinities for Detection of Pancreatic Cancer" Clin. Cancer Res. 2012 18:839–849.
|
|
Pirie, C.M., Hackel, B.J., Rosenblum, M.G., and Wittrup, K.D., "Convergent potency
of internalized gelonin immunotoxins across varied cell lines, antigens, and targeting
moieties" J. Biol. Chem. 2011 286, 4165-4172.
|
|
Hackel, B.J., Ackerman, M., Howland, S., and Wittrup, K.D., "Stability and
complementarity-determining region biases enrich binder functionality landscapes"
J. Mol. Biol. 2010 401, 84-96.
|
|
Hackel, B.J., and Wittrup, K.D., "The full amino acid repertoire is superior to
serine/tyrosine for selection of high affinity immunoglobulin G binders from the
fibronectin scaffold" Prot. Engr. Des. Sel. 2010 23, 211-219.
|
|
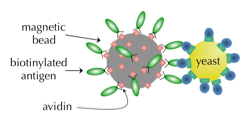
Ackerman, M., Levary, D., Tobon, G., Hackel, B.J., Orcutt, K.D., and Wittrup, K.D.,
"Highly avid magnetic bead capture: an efficient selection method for de novo protein
engineering utilizing yeast surface display" Biotech. Prog. 2009 25, 774-783.
|
|
Hackel, B.J., Kapila, A., and Wittrup, K.D., "Picomolar affinity fibronectin domains
engineered utilizing loop length diversity, recursive mutagenesis, and loop shuffling"
J. Mol. Biol. 2008 381, 1236-1252.
|
|
Lipovsek, D., Lippow, S.M., Hackel, B.J., Gregson, M.W., Cheng, P., Kapila, A., and
Wittrup, K.D., "Evolution of an interloop disulfide bond in high-affinity antibody
mimics based on fibronectin type III domain and selected by yeast-surface display:
Molecular convergence with single-domain camelid and shark antibodies"
J. Mol. Biol. 2007 368, 1024-1041.
|
|
Chao, G., Lau, W.L., Hackel, B.J., Sazinsky, S.L., Lippow, S.M., and Wittrup, K.D.,
"Isolating and engineering human antibodies using yeast surface display"
Nature Protocols 2006 1, 755-768.
|
|
Hackel, B.J., Huang, D., Bubolz, J.C., Wang, X.X., and Shusta, E.V.,
"Production of soluble and active transferrin receptor-targeting single-chain
antibody using Saccharomyces cerevisiae" Pharm. Res. 2006 23, 790-797.
|
|
Camarero J.A., Hackel, B.J., DeYoreo, J.J, and Mitchell, A.R., "Fmoc-based synthesis
of peptide alpha-thioesters using an aryl hydrazine support" J. Org. Chem. 2004 69, 4145-51.
|
|
|
|
Hackel, B.J., "Alternative Protein Scaffolds for Molecular Imaging and Therapy"
in Engineering in Translational Medicine (Cai, W., ed) 2014, 343-364, Springer.
|

|
Hackel, B.J., and Wittrup, K.D., "Yeast surface display in protein engineering and analysis" in
Protein Engineering Handbook (Lutz, S., and Bornscheuer, U.T., eds) 2008, 621-648. Wiley.
|
Patents
|

|
Hackel, B.J., Kruziki, M.A., Bhatnagar, S., and Zhou, H., "Protein scaffolds and methods of use."
PCT/US2014/063441; October 31, 2014
|
Software
|

|
PACT: Protein Analysis and Classifier Toolkit
|

|
PyMOL360: Multi-user gamepad control of molecular visualization software
|

|
ScaffoldSeq: Software for characterization of directed evolution populations (ScaffoldSeq.zip)
|